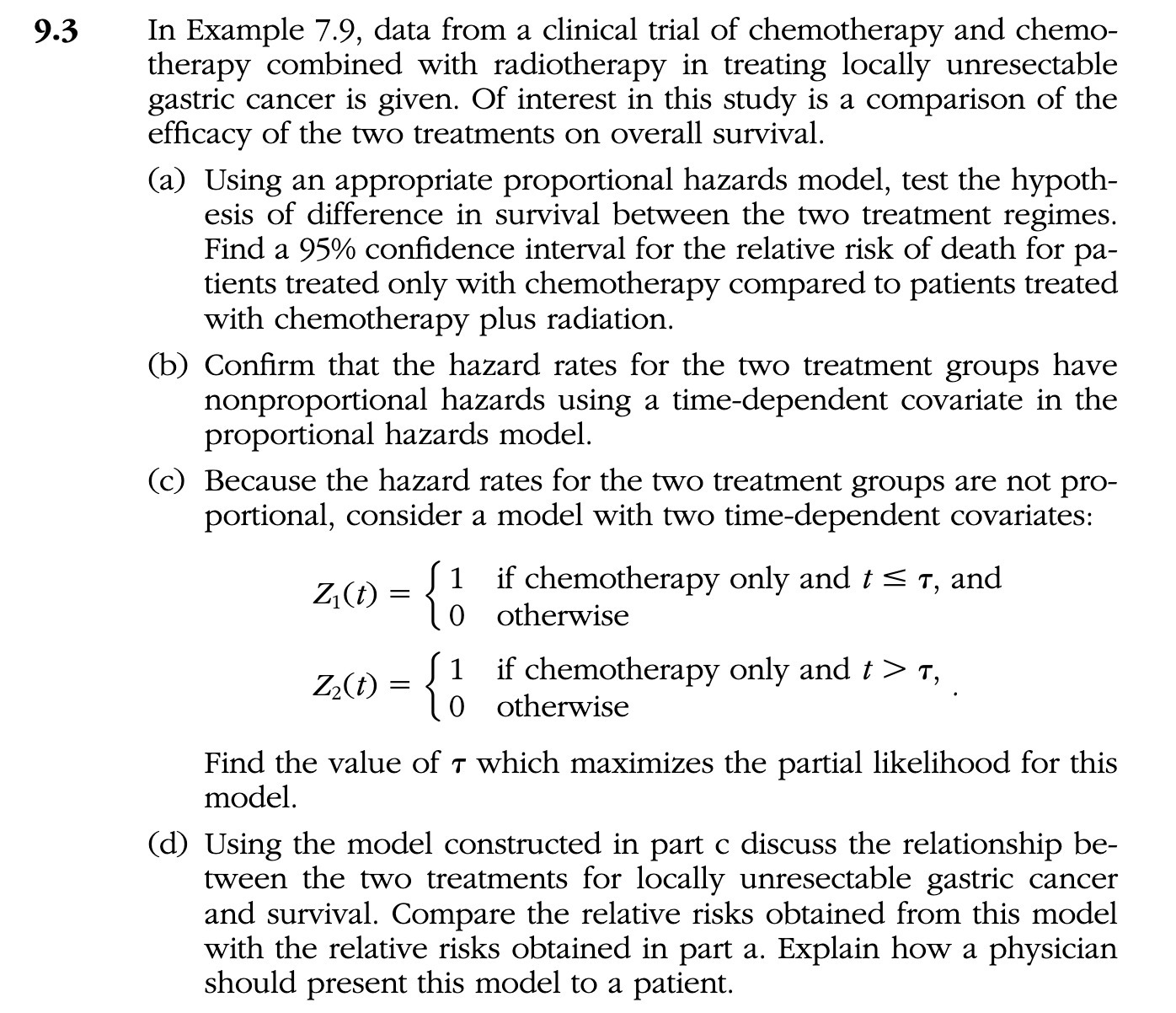
library(survival)
library(ClinicalTrialSummary)
data(ggas)
# group 0: Chemotherapy only; 1: Chemotherapy plus radiotherapy
dat <- ggas
dat$time <- dat$time * 365
dat$therapy <- ifelse(dat$group == 0, 1, 0)
model_a <- coxph(Surv(time, event) ~ therapy, dat)
summary(model_a)
Call:
coxph(formula = Surv(time, event) ~ therapy, data = dat)
n= 90, number of events= 82
coef exp(coef) se(coef) z Pr(>|z|)
therapy -0.1051 0.9002 0.2233 -0.471 0.638
exp(coef) exp(-coef) lower .95 upper .95
therapy 0.9002 1.111 0.5811 1.395
Concordance= 0.562 (se = 0.031 )
Likelihood ratio test= 0.22 on 1 df, p=0.6
Wald test = 0.22 on 1 df, p=0.6
Score (logrank) test = 0.22 on 1 df, p=0.6
model_b <- coxph(Surv(time, event) ~ therapy + tt(therapy), dat,
tt = function(x, t, ...)x*log(t))
summary(model_b)
Call:
coxph(formula = Surv(time, event) ~ therapy + tt(therapy), data = dat,
tt = function(x, t, ...) x * log(t))
n= 90, number of events= 82
coef exp(coef) se(coef) z Pr(>|z|)
therapy -3.86619 0.02094 1.47707 -2.617 0.00886 **
tt(therapy) 0.64677 1.90935 0.24793 2.609 0.00909 **
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
exp(coef) exp(-coef) lower .95 upper .95
therapy 0.02094 47.7602 0.001158 0.3786
tt(therapy) 1.90935 0.5237 1.174488 3.1040
Concordance= 0.576 (se = 0.03 )
Likelihood ratio test= 8.54 on 2 df, p=0.01
Wald test = 6.87 on 2 df, p=0.03
Score (logrank) test = 7.59 on 2 df, p=0.02
tao <- unique(sort(dat$time[dat$event==1]))
l <- numeric(length(tao))
for (i in 1:length(tao)) {
temp <- survSplit(Surv(time, event) ~ group, data= dat, cut=c(tao[i]),
episode= "tgroup", id="id")
fit <- coxph(Surv(tstart, time, event) ~ group +
group:factor(tgroup), data=temp)
l[i] <- logLik(fit)
}
Warning in agreg.fit(X, Y, istrat, offset, init, control, weights = weights, :
Loglik converged before variable 1,2 ; beta may be infinite.
Warning in agreg.fit(X, Y, istrat, offset, init, control, weights = weights, :
Loglik converged before variable 2 ; beta may be infinite.
(taoHat <- tao[which.max(l)])
temp_d <- survSplit(Surv(time, event) ~ group, data= dat, cut=254,
episode= "tgroup", id="id")
temp_d$group1 <- ifelse(temp_d$group==0 & temp_d$tgroup==1,1,0)
temp_d$group2 <- ifelse(temp_d$group==0 & temp_d$tgroup==2,1,0)
fit_d2 <- coxph(Surv(tstart, time, event) ~ group1 + group2, data=temp_d)
summary(fit_d2)
Call:
coxph(formula = Surv(tstart, time, event) ~ group1 + group2,
data = temp_d)
n= 150, number of events= 82
coef exp(coef) se(coef) z Pr(>|z|)
group1 -1.4229 0.2410 0.4327 -3.288 0.00101 **
group2 0.6423 1.9008 0.3048 2.107 0.03508 *
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
exp(coef) exp(-coef) lower .95 upper .95
group1 0.241 4.1491 0.1032 0.5628
group2 1.901 0.5261 1.0460 3.4542
Concordance= 0.62 (se = 0.029 )
Likelihood ratio test= 17.79 on 2 df, p=1e-04
Wald test = 15.25 on 2 df, p=5e-04
Score (logrank) test = 17.31 on 2 df, p=2e-04